=== Training HMM with K=2 states ===
0 ELBO = 190,232
10 ELBO = 193,415
20 ELBO = 159,915
30 ELBO = 375,032
40 ELBO = 175,100
50 ELBO = 210,764
60 ELBO = 178,541
70 ELBO = 172,792
80 ELBO = 236,583
90 ELBO = 227,445
100 ELBO = 316,513
110 ELBO = 184,105
120 ELBO = 171,063
130 ELBO = 203,254
140 ELBO = 186,968
150 ELBO = 192,389
160 ELBO = 203,597
170 ELBO = 174,310
180 ELBO = 202,074
190 ELBO = 165,841
200 ELBO = 172,105
210 ELBO = 206,842
220 ELBO = 178,325
230 ELBO = 178,878
240 ELBO = 228,023
250 ELBO = 206,081
260 ELBO = 222,080
270 ELBO = 153,061
280 ELBO = 204,136
290 ELBO = 166,776
300 ELBO = 197,441
310 ELBO = 166,890
320 ELBO = 167,722
330 ELBO = 194,650
340 ELBO = 180,926
350 ELBO = 185,720
360 ELBO = 176,313
370 ELBO = 194,673
380 ELBO = 162,984
390 ELBO = 167,645
400 ELBO = 142,858
410 ELBO = 284,911
420 ELBO = 168,510
430 ELBO = 171,370
440 ELBO = 283,396
450 ELBO = 172,392
460 ELBO = 205,640
470 ELBO = 219,136
480 ELBO = 198,483
490 ELBO = 163,924
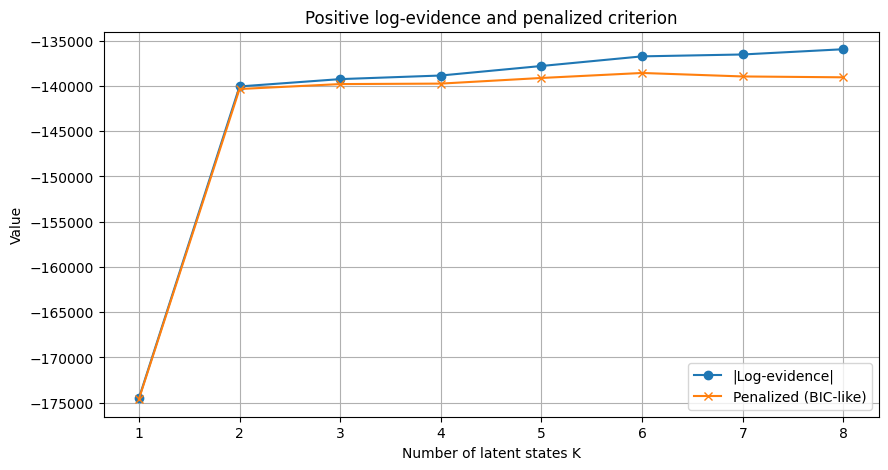
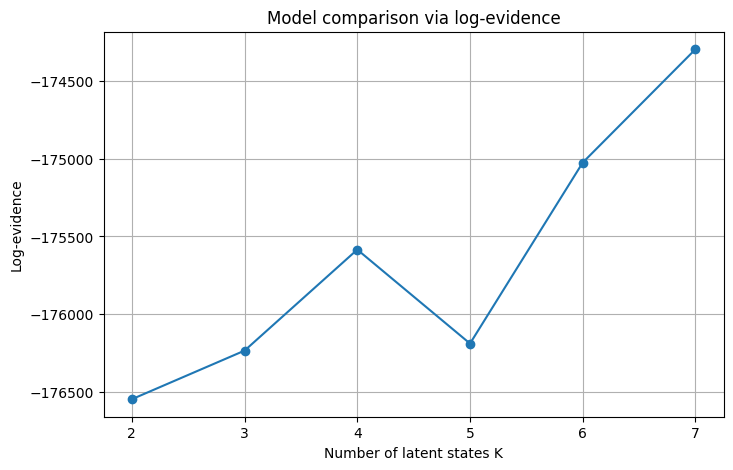
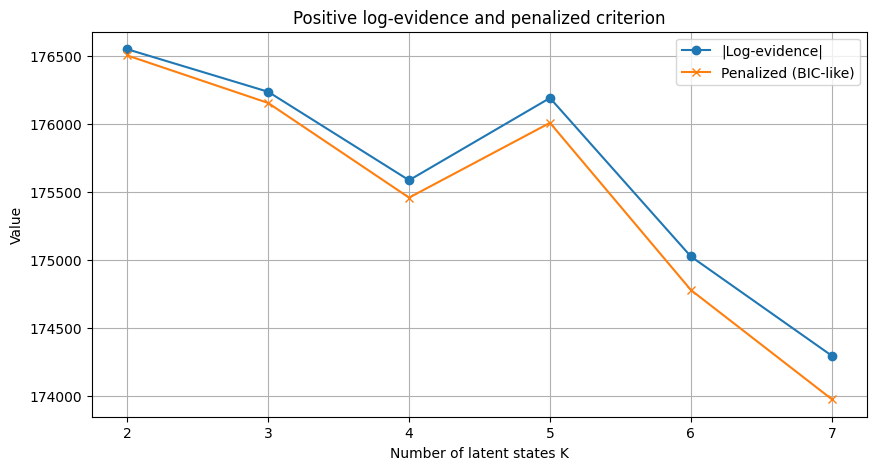
Log-evidence K=2: -176548.53
=== Training HMM with K=3 states ===
0 ELBO = 247,371
10 ELBO = 172,018
20 ELBO = 193,899
30 ELBO = 227,453
40 ELBO = 191,811
50 ELBO = 298,896
60 ELBO = 200,825
70 ELBO = 178,750
80 ELBO = 199,435
90 ELBO = 178,607
100 ELBO = 243,646
110 ELBO = 178,217
120 ELBO = 182,281
130 ELBO = 172,379
140 ELBO = 166,560
150 ELBO = 182,253
160 ELBO = 267,153
170 ELBO = 220,679
180 ELBO = 178,813
190 ELBO = 200,628
200 ELBO = 175,836
210 ELBO = 178,554
220 ELBO = 181,986
230 ELBO = 172,293
240 ELBO = 173,313
250 ELBO = 178,147
260 ELBO = 173,142
270 ELBO = 198,315
280 ELBO = 166,034
290 ELBO = 186,686
300 ELBO = 218,927
310 ELBO = 177,696
320 ELBO = 173,187
330 ELBO = 173,021
340 ELBO = 169,031
350 ELBO = 184,694
360 ELBO = 173,284
370 ELBO = 174,705
380 ELBO = 169,267
390 ELBO = 165,636
400 ELBO = 190,386
410 ELBO = 178,986
420 ELBO = 173,631
430 ELBO = 171,541
440 ELBO = 168,006
450 ELBO = 166,954
460 ELBO = 147,162
470 ELBO = 163,471
480 ELBO = 154,490
490 ELBO = 190,566
Log-evidence K=3: -176235.47
=== Training HMM with K=4 states ===
0 ELBO = 240,320
10 ELBO = 312,726
20 ELBO = 171,498
30 ELBO = 185,044
40 ELBO = 210,946
50 ELBO = 215,290
60 ELBO = 196,785
70 ELBO = 251,274
80 ELBO = 235,890
90 ELBO = 167,417
100 ELBO = 181,020
110 ELBO = 200,026
120 ELBO = 193,995
130 ELBO = 173,299
140 ELBO = 179,895
150 ELBO = 161,756
160 ELBO = 161,841
170 ELBO = 172,566
180 ELBO = 274,967
190 ELBO = 186,367
200 ELBO = 177,715
210 ELBO = 176,197
220 ELBO = 167,204
230 ELBO = 193,153
240 ELBO = 215,687
250 ELBO = 176,218
260 ELBO = 171,713
270 ELBO = 165,249
280 ELBO = 161,930
290 ELBO = 166,931
300 ELBO = 167,066
310 ELBO = 172,889
320 ELBO = 177,175
330 ELBO = 175,511
340 ELBO = 175,998
350 ELBO = 184,089
360 ELBO = 190,777
370 ELBO = 162,425
380 ELBO = 168,862
390 ELBO = 177,483
400 ELBO = 183,929
410 ELBO = 182,565
420 ELBO = 168,867
430 ELBO = 187,491
440 ELBO = 204,556
450 ELBO = 155,868
460 ELBO = 180,376
470 ELBO = 229,833
480 ELBO = 171,871
490 ELBO = 184,568
Log-evidence K=4: -175584.80
=== Training HMM with K=5 states ===
0 ELBO = 301,775
10 ELBO = 265,383
20 ELBO = 168,610
30 ELBO = 196,518
40 ELBO = 268,005
50 ELBO = 287,431
60 ELBO = 181,713
70 ELBO = 208,820
80 ELBO = 173,086
90 ELBO = 200,453
100 ELBO = 164,948
110 ELBO = 230,225
120 ELBO = 189,927
130 ELBO = 180,045
140 ELBO = 188,618
150 ELBO = 205,706
160 ELBO = 174,525
170 ELBO = 170,300
180 ELBO = 234,548
190 ELBO = 174,461
200 ELBO = 183,315
210 ELBO = 188,289
220 ELBO = 174,339
230 ELBO = 167,151
240 ELBO = 182,117
250 ELBO = 205,801
260 ELBO = 161,744
270 ELBO = 177,010
280 ELBO = 166,169
290 ELBO = 188,263
300 ELBO = 177,360
310 ELBO = 173,780
320 ELBO = 157,469
330 ELBO = 172,907
340 ELBO = 174,215
350 ELBO = 199,236
360 ELBO = 178,875
370 ELBO = 166,361
380 ELBO = 171,062
390 ELBO = 164,785
400 ELBO = 184,789
410 ELBO = 188,330
420 ELBO = 173,451
430 ELBO = 170,524
440 ELBO = 158,213
450 ELBO = 160,903
460 ELBO = 169,181
470 ELBO = 167,878
480 ELBO = 190,540
490 ELBO = 181,442
Log-evidence K=5: -176189.83
=== Training HMM with K=6 states ===
0 ELBO = 205,246
10 ELBO = 204,992
20 ELBO = 178,281
30 ELBO = 224,585
40 ELBO = 184,819
50 ELBO = 193,440
60 ELBO = 262,086
70 ELBO = 193,501
80 ELBO = 179,400
90 ELBO = 193,507
100 ELBO = 165,328
110 ELBO = 167,362
120 ELBO = 191,785
130 ELBO = 196,277
140 ELBO = 177,968
150 ELBO = 201,942
160 ELBO = 168,274
170 ELBO = 177,236
180 ELBO = 197,133
190 ELBO = 170,824
200 ELBO = 170,287
210 ELBO = 170,944
220 ELBO = 168,661
230 ELBO = 167,795
240 ELBO = 174,382
250 ELBO = 166,777
260 ELBO = 186,454
270 ELBO = 151,819
280 ELBO = 167,925
290 ELBO = 169,870
300 ELBO = 174,078
310 ELBO = 171,096
320 ELBO = 169,600
330 ELBO = 171,617
340 ELBO = 170,151
350 ELBO = 175,450
360 ELBO = 172,908
370 ELBO = 166,262
380 ELBO = 170,416
390 ELBO = 171,710
400 ELBO = 164,363
410 ELBO = 178,430
420 ELBO = 172,183
430 ELBO = 155,204
440 ELBO = 170,694
450 ELBO = 170,051
460 ELBO = 158,892
470 ELBO = 208,772
480 ELBO = 168,031
490 ELBO = 174,782
Log-evidence K=6: -175024.06
=== Training HMM with K=7 states ===
0 ELBO = 219,438
10 ELBO = 196,574
20 ELBO = 202,997
30 ELBO = 194,856
40 ELBO = 270,244
50 ELBO = 241,503
60 ELBO = 214,216
70 ELBO = 172,002
80 ELBO = 172,319
90 ELBO = 177,734
100 ELBO = 170,084
110 ELBO = 262,432
120 ELBO = 190,945
130 ELBO = 211,321
140 ELBO = 166,894
150 ELBO = 210,115
160 ELBO = 187,824
170 ELBO = 173,195
180 ELBO = 173,743
190 ELBO = 205,510
200 ELBO = 189,702
210 ELBO = 200,941
220 ELBO = 181,681
230 ELBO = 218,355
240 ELBO = 173,864
250 ELBO = 170,276
260 ELBO = 171,114
270 ELBO = 174,103
280 ELBO = 185,191
290 ELBO = 164,321
300 ELBO = 193,377
310 ELBO = 177,413
320 ELBO = 181,125
330 ELBO = 166,970
340 ELBO = 179,617
350 ELBO = 178,753
360 ELBO = 177,921
370 ELBO = 166,222
380 ELBO = 174,462
390 ELBO = 163,592
400 ELBO = 178,974
410 ELBO = 174,073
420 ELBO = 164,714
430 ELBO = 179,232
440 ELBO = 165,262
450 ELBO = 177,316
460 ELBO = 169,859
470 ELBO = 176,121
480 ELBO = 166,555
490 ELBO = 168,900
Log-evidence K=7: -174293.88